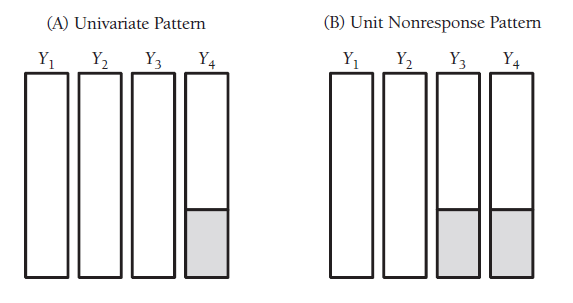
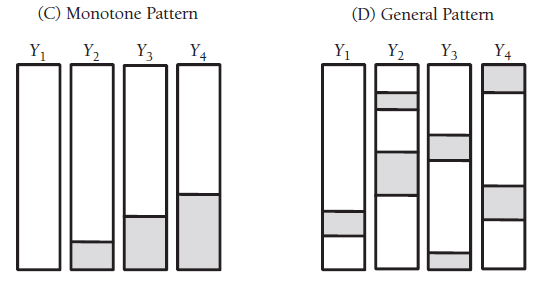
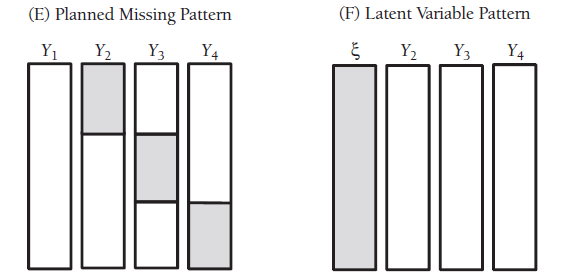
frause wage2, clear
set seed 312
xtile smp = runiform()
replace smp=smp==1
gen wage_s = wage if smp==1
**three Matching scores
** Pmm
reg wage_s hours iq kww educ exper tenure age married black south urban sibs
predict wageh
** pscore
logit smp hours iq kww educ exper tenure age married black south urban sibs
predict pscore, xb
** pca
pca hours iq kww educ exper tenure age married black south urban sibs , comp(1)
predict pc1
foreach i in wageh pscore pc1 {
qui:sum `i'
replace `i' = (`i'-r(mean))/r(sd)
}(467 real changes made)
(467 missing values generated)
Source | SS df MS Number of obs = 468
-------------+---------------------------------- F(12, 455) = 14.83
Model | 24043666.1 12 2003638.84 Prob > F = 0.0000
Residual | 61493527.5 455 135150.61 R-squared = 0.2811
-------------+---------------------------------- Adj R-squared = 0.2621
Total | 85537193.6 467 183163.155 Root MSE = 367.63
------------------------------------------------------------------------------
wage_s | Coefficient Std. err. t P>|t| [95% conf. interval]
-------------+----------------------------------------------------------------
hours | -4.080234 2.204689 -1.85 0.065 -8.412869 .2524015
iq | 3.090831 1.459171 2.12 0.035 .2232806 5.958381
kww | 5.637987 2.888128 1.95 0.052 -.0377362 11.31371
educ | 53.43222 10.41882 5.13 0.000 32.95724 73.9072
exper | 8.816439 5.30953 1.66 0.098 -1.617804 19.25068
tenure | 6.327233 3.534248 1.79 0.074 -.6182417 13.27271
age | 10.92113 7.195943 1.52 0.130 -3.220278 25.06253
married | 143.306 54.35023 2.64 0.009 36.49735 250.1146
black | -144.0597 60.51784 -2.38 0.018 -262.9889 -25.1306
south | -37.37316 37.7745 -0.99 0.323 -111.6073 36.86096
urban | 200.3258 38.93558 5.15 0.000 123.81 276.8417
sibs | 1.881618 8.468635 0.22 0.824 -14.76087 18.52411
_cons | -842.6706 273.28 -3.08 0.002 -1379.718 -305.6231
------------------------------------------------------------------------------
(option xb assumed; fitted values)
Iteration 0: Log likelihood = -648.09208
Iteration 1: Log likelihood = -642.21625
Iteration 2: Log likelihood = -642.21522
Iteration 3: Log likelihood = -642.21522
Logistic regression Number of obs = 935
LR chi2(12) = 11.75
Prob > chi2 = 0.4657
Log likelihood = -642.21522 Pseudo R2 = 0.0091
------------------------------------------------------------------------------
smp | Coefficient Std. err. z P>|z| [95% conf. interval]
-------------+----------------------------------------------------------------
hours | .0153544 .0093201 1.65 0.099 -.0029127 .0336215
iq | .0024677 .0057346 0.43 0.667 -.0087719 .0137072
kww | -.0095774 .0113247 -0.85 0.398 -.0317735 .0126186
educ | -.0255069 .0409702 -0.62 0.534 -.105807 .0547932
exper | .0256009 .0205087 1.25 0.212 -.0145955 .0657973
tenure | .0131078 .0137404 0.95 0.340 -.013823 .0400385
age | -.0033883 .0281317 -0.12 0.904 -.0585256 .0517489
married | -.2382151 .2167632 -1.10 0.272 -.6630632 .1866331
black | -.1214461 .2276112 -0.53 0.594 -.5675559 .3246637
south | -.0370746 .1457381 -0.25 0.799 -.3227161 .2485669
urban | -.0371736 .1495076 -0.25 0.804 -.3302031 .255856
sibs | -.041735 .0313008 -1.33 0.182 -.1030835 .0196135
_cons | -.1246922 1.056515 -0.12 0.906 -2.195424 1.946039
------------------------------------------------------------------------------
Principal components/correlation Number of obs = 935
Number of comp. = 1
Trace = 12
Rotation: (unrotated = principal) Rho = 0.2112
--------------------------------------------------------------------------
Component | Eigenvalue Difference Proportion Cumulative
-------------+------------------------------------------------------------
Comp1 | 2.5348 .622213 0.2112 0.2112
Comp2 | 1.91258 .821455 0.1594 0.3706
Comp3 | 1.09113 .0297471 0.0909 0.4615
Comp4 | 1.06138 .0497587 0.0884 0.5500
Comp5 | 1.01162 .0867533 0.0843 0.6343
Comp6 | .924871 .0639678 0.0771 0.7114
Comp7 | .860903 .0907871 0.0717 0.7831
Comp8 | .770116 .144885 0.0642 0.8473
Comp9 | .625231 .119791 0.0521 0.8994
Comp10 | .50544 .0919858 0.0421 0.9415
Comp11 | .413454 .124988 0.0345 0.9760
Comp12 | .288466 . 0.0240 1.0000
--------------------------------------------------------------------------
Principal components (eigenvectors)
--------------------------------------
Variable | Comp1 | Unexplained
-------------+----------+-------------
hours | 0.1320 | .9558
iq | 0.4921 | .3863
kww | 0.4220 | .5486
educ | 0.4555 | .474
exper | -0.2161 | .8817
tenure | 0.0463 | .9946
age | 0.0527 | .993
married | 0.0053 | .9999
black | -0.3722 | .6488
south | -0.2076 | .8908
urban | 0.0723 | .9868
sibs | -0.3411 | .7051
--------------------------------------
(score assumed)
Scoring coefficients
sum of squares(column-loading) = 1
------------------------
Variable | Comp1
-------------+----------
hours | 0.1320
iq | 0.4921
kww | 0.4220
educ | 0.4555
exper | -0.2161
tenure | 0.0463
age | 0.0527
married | 0.0053
black | -0.3722
south | -0.2076
urban | 0.0723
sibs | -0.3411
------------------------
(935 real changes made)
(935 real changes made)
(935 real changes made)